FindingPheno develops new tools to disentangle biological interactions between host and microbiomes
The FindingPheno project will improve how we understand and utilise the functions provided by microbiomes in combating human diseases as well as the way we produce sustainable food for future generations.
What is the aim of FindingPheno?
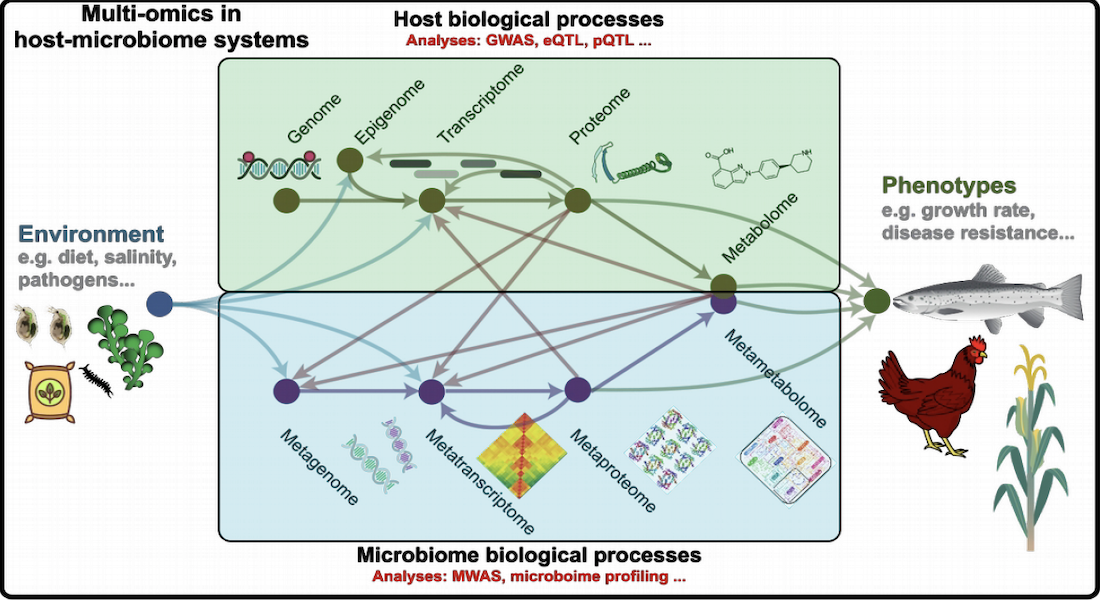
Multi-omics data, illustrated in the figure above, is becoming the norm in genomics studies, with data from microbiomes becoming an integral part of the standard complement collected and sequenced in genomics experiments. But methods that allow an integrated analysis of the multi-omics data are still lacking, resulting in diminished understanding of host-microbiome interactions. To that end, FindingPheno will develop computational tools to decipher biomolecular interactions between hosts and microbiomes by combining biological knowledge and state-of-the-art statistical methods. The primary aim of this action is to develop and demonstrate a novel computational platform to analyse multi-omics data to identify the complex molecular basis of organismal phenotypes.
Why is this project important?
“The explosion of research into the functions provided by associated microbiomes to their host organism is currently generating big multi-omics data sets at an unprecedented rate. This accumulation of BIG holistic data sets is bringing with it an urgent need for the development of new computational methods to better harvest information on the truly interesting interaction processes by which host organisms and their microbiomes interact to shape the ultimate phenotypes of e.g. disease resistance and growth efficiency. FindingPheno addresses this exact knowledge gap by using existing BIG data sets to develop new methods to better analyse these data; the methods developed in FindingPheno are expected to significantly improve how we understand and utilise the functions provided by microbiomes in combating human diseases as well as the way we produce sustainable food for future generations.” Co-PI and Assoc Prof Morten Limborg says.
Applied studies will ensure the data
This framework will be applied to case studies from actual food production systems, using unique multi-omics data sets from chicken, salmon, tomatoes and bees. Applied studies such as the HoloFood project, that are already being carried out at the Center for Evolutionary Hologenomics will assure the data. We expect to show how to improve the effectiveness of microbiome interventions in sustainable food production, and simultaneously, we will offer avenues for quick and easy application of this new approach to other relevant biotechnology-based industries, e.g. enzyme production and fermentation.
Built around an international consortium
FindingPheno was granted 5.8 million € in total and 13.7 million DKK will go to University of Copenhagen. The project is built around an international consortium of 8 partners including 5 Universities/Research institutions (University of Copenhagen (Denmark), University of Turku (Finland), Center for Ecological Research (Hungary), EMBL/EBI (Germany/UK) and Champalimaud Foundation (Portugal)) as well as 3 Private enterprises (Chr. Hansen A/S (Denmark), Qiagen A/S (Denmark) and Njorth Bio Science Ltd. (UK)) that will devise novel computational methods to infer the interactions between the host genomics and the gut microbial communities using multi-omics data, to better understand the role of this interaction on systems ranging from salmon, chicken, honey bees, maize, and tomato.
In the video below you can watch coordinator and Assoc Prof Shyam Gopalakrishnan explain more about the project.
What was your motivation for applying for this grant?
“I had unsuccessfully applied for similar grants at the Danish national level, that were designed to be a single institution project, but basically doing a subset of the same things that we propose to do in FindingPheno. When I saw that "genotype-phenotype association" was the cornerstone of this Research and Innovation Action topic, a light went off in my head that we needed to extend the smaller projects to answer bigger open questions involving the integration of multi-omics datasets - with a special focus on microbiomes and their interactions with host genomes, to better understand the role these interactions play in different phenotypes.” Coordinator and Assoc Prof Shyam Gopalakrishnan says.
Who helped you during the writing process?
Shyam continues: “Morten (Limborg) and I brought different skills to the table for the grant writing process, where I focused on the excellence and the implementation sections, and Morten led the impact section, and helped coalesce the application into a coherent whole. Our liaison at the EU pre-award office - Astrid Cermak - was absolutely fantastic to work with during the grant writing process for both the first and second stage of the application. She not only acted as the expert in the structure and language of such grant, but was also as a wonderful colleague helping us balance the excellence and impact sections of the application. ”
What’s next?
Contacts


