We combine whole genomes, archaeological evidence and traditional knowledge to learn about the evolutionary history of Indigenous Americans.
Reconstructing demographic histories in the Ancient Population Genomics Group
The Ancient Population Genomics Group generates present-day and ancient population genomic data and contextualises them with archaeological evidence and traditional knowledge to reconstruct the evolutionary history of human populations.
Our research involves population genomics, ancient DNA, human evolution and computational biology.
By sequencing DNA retrieved from present-day and ancient individuals, we reconstruct human population history. Furthermore, we use DNA retrieved from perishable archaeological materials to understand not only the demographic history of human populations but also how it was shaped by environmental factors, e.g., diet, disease and cultural innovations and exchange with other groups. Since our work is driven by biological, archaeological and anthropological questions, we collaborate with researchers specialising in diverse disciplines to interpret our results jointly with archaeological evidence and traditional knowledge.
Although we have a broad interest in human populations worldwide, we focus our research on Indigenous American population history and three major historical transitions: the early quick peopling by ice age hunter gatherers; the rise of Holocene complex agricultural societies; and their decline due to European colonisation.
In addition, our research has contributed biological data that questions widespread colonial narratives such as the "Palaeoamerican Model" for the peopling of the Americas and the alleged population "collapse" on Rapa Nui.
Student projects are always available, so please write us if you are interested in joining!
- Early human dispersals within the Americas. Moreno-Mayar JV*, Vinner L*, de Barros Damgaard P*, de la Fuente C*, Chan J*, Spence JP*, [44 authors], Nielsen R, Song YS, Meltzer DJ, Willerslev E. Science. 2018; 362(6419): eaav2621. doi: 10.1126/science.aav2621
- Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans. Moreno-Mayar JV*, Potter BA*, Vinner L*, [11 authors], Song YS, Nielsen R, Meltzer DJ, Willerslev E. Nature. 2018; 553(7687): 203-207. doi: 10.1038/nature25173
- Ancient Rapanui genomes reveal resilience and pre-European contact with the Americas. Moreno-Mayar JV*‡, Sousa da Mota B*, [12 authors], Malaspinas AS‡. Nature. 2024; 633(8029): 389-397. doi: 10.1038/s41586-024-07881-4
- A likelihood method for estimating present-day human contamination in ancient male samples using low-depth X- chromosome data. Moreno-Mayar JV*‡, Korneliussen TS*,Dalal J, Renaud G, Albrechtsen A, Nielsen R, Malaspinas AS‡. Bioinformatics. 2019; btz660. doi: 10.1093/bioinformatics/btz660
- FrAnTK: a Frequency-based Analysis ToolKit for efficient exploration of allele sharing patterns in present-day and ancient genomic datasets. Moreno-Mayar JV‡. G3 (Bethesda). 2021; 12(1): jkab357. doi:10.1093/g3journal/jkab357
Full list of publications by J. Víctor Moreno-Mayar
Projects
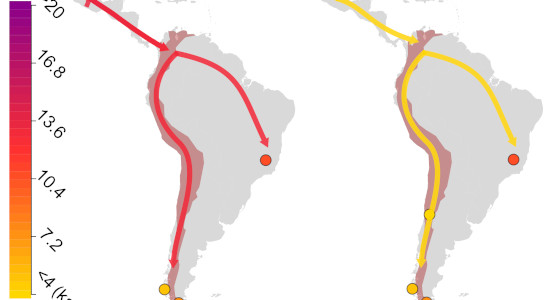
Peopling of the Americas
Quids
We retrieve DNA from masticated plants to learn about the evolutionary history of the chewers, their diet, their oral microbiome and their interplay.

Peopling of Polynesia
We combine whole genomes, archaeological evidence and traditional knowledge to learn about the population history of Polynesians.

Bioinformatics methods
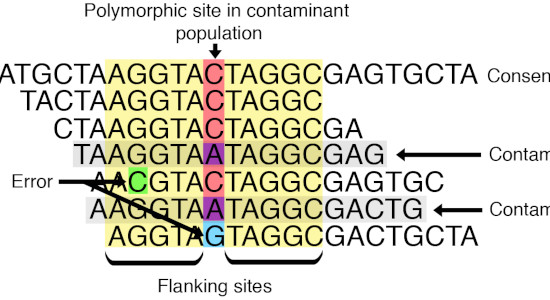
We develop bioinformatic methods that take into account contamination and post-mortem damage in ancient DNA genome datasets.

Group Leader
J. Víctor Moreno-Mayar
Assistant Professor
Phone: +45 35337807
E-mail: morenomayar@sund.ku.dk
Group members
| Name | Title | Phone | |
|---|---|---|---|
| Auguste Kontvainaite | Visitor | ||
| Lorraine Helyett Irène Bocquet-Appel | Guest Researcher | ||
| Sabrina Michelle Schum | Visiting Student | ||
| Signe Klemm | PhD Fellow | +4535335128 | |
| Sofía Ivonne Vieyra Sánchez | PhD Fellow | +4535329218 |



