Reconstructing ancient plant populations using fossil pollen DNA
Conservation Palaeogenetics & Palaeoecology Group (COPAL)
The Conservation Palaeogenetics & Palaeoecology Group seeks to generate knowledge, methods, and tools relevant to resolving some of the key challenges in biodiversity conservation, including climate change, land use change and emerging diseases. We work at the interface of ancient genetics, palaeoecology, evolutionary biology, and conservation science to understand how species and ecosystems responded to past environmental change and how this understanding can inform conservation decision-making.

We are amid the sixth mass extinction and funds for combating it are limited. Consequently, there is a pressing need to maintain and enhance the resilience of species and ecosystems to environmental change. However, much of the genetic diversity underpinning this resilience has been lost over the past decades, centuries, and millennia due to human impacts. Ancient genomics allows us to track biological responses to past climatic and anthropogenic changes, from genes to populations to ecological communities. This approach represents a powerful new way to recover ‘lost’ genetic diversity relevant to preserving Earth’s contemporary biodiversity into the future.
Our research centres around two questions: What is the genetic basis of resilience to environmental stressors, and how can it be used to guide conservation and restoration of biodiversity? To answer these questions, we use ancient genomic methods in tandem with other palaeo techniques, experimental approaches, and observational data, in close collaboration with academic colleagues and other partners from across the globe. The overarching aim is to uncover genetic underpinning of favourable phenotypic responses to the environmental stressors projected to amplify in the coming decades. To maximise the impact of our research efforts, we focus on threatened keystone species from globally important ecosystems.
Sand, K. K., Jelavić, S., Kjær, K. H., & Prohaska, A. (2024). Importance of eDNA taphonomy and sediment provenance for robust ecological inference: Insights from interfacial geochemistry. Environmental DNA, 6, e519.
Prohaska A., Seddon A.W.R., Smith A., Rach O., Sachse D., and K.J. Willis. Long-Term Ecological Responses of a Dipterocarp Forest to Climate Changes and Nutrient Availability. New Phytologist 240(6):2513-2529 (2023). *Corresponding author
Prohaska, A.*, Seddon, A.W.R., Meese, B., Chiang, J.C.H., Willis, K.J., Sachse, D. Abrupt change in tropical Pacific climate mean state during the Little Ice Age. Communications Earth & Environment 4, 227 (2023). *Corresponding author
Wang, Y., Pedersen, M. W., Alsos, I. G., et al. (2021) Late Quaternary dynamics of Arctic biota from ancient environmental genomics. Nature, 1-7. *These authors contributed equally to this work.
Grace M.K., et al. (2021) Testing a global standard for quantifying species recovery and assessing conservation impact. Conservation Biology Conservation Biology, 1–17.
Prohaska A.*, Racimo A.*, Schork A. J.*, et al. (2019) Human disease variation in the light of population genomics. Cell 177, pp. 115–31. *These authors contributed equally to this work.
Cappellini E.*, Prohaska A.*, et al. (2018) Ancient biomolecules and evolutionary inference. Annual Review of Biochemistry 87, pp. 1029-1060. *These authors contributed equally to this work.
Projects
THE FOSSIL POLLEN SEQUENCING PROJECT
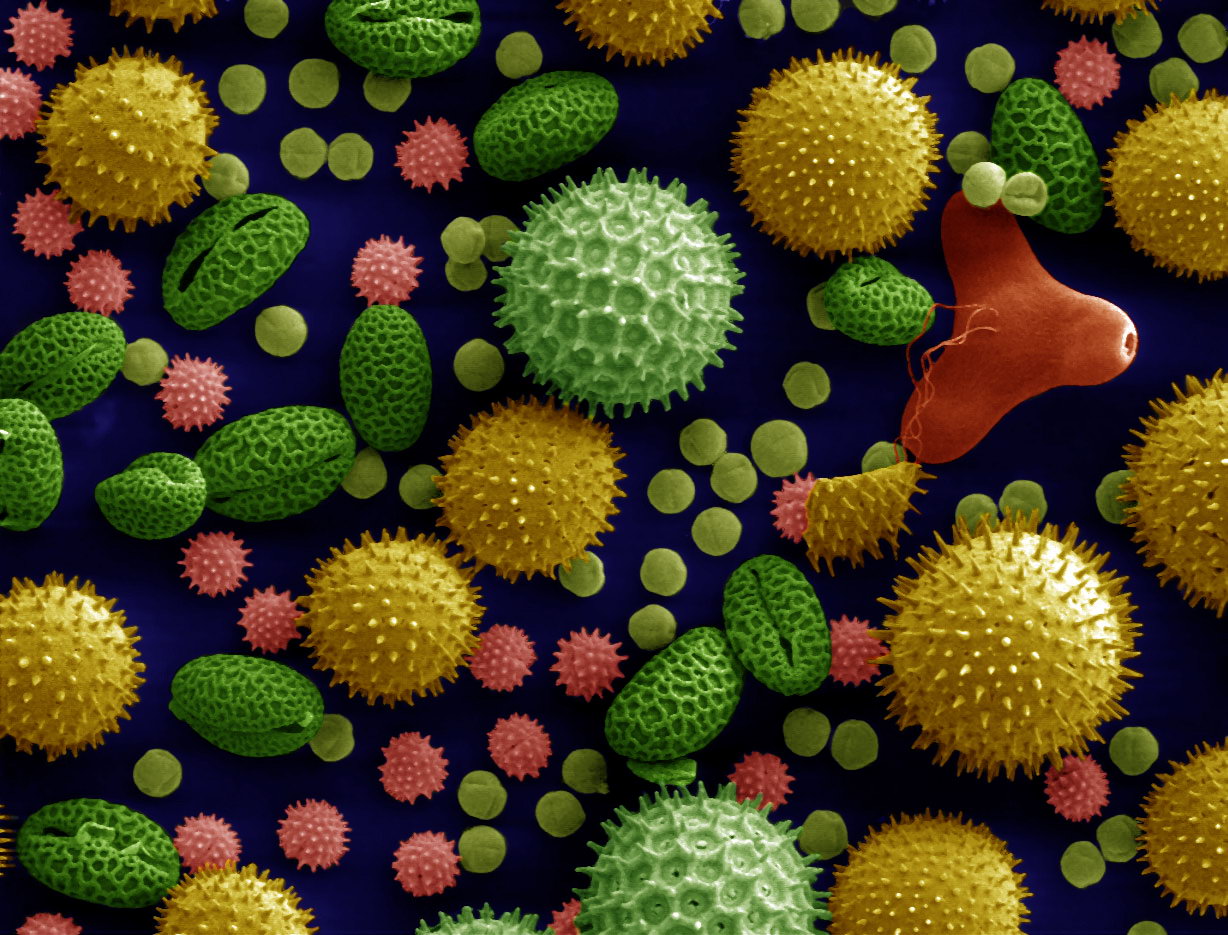
The Ancient Coral Genomics Project
Reconstructing coral reef response to past climate changes using ancient DNA

The Great Adriatic Plain Project
Reconstructing plant and animal assemblages during the Last Glacial using ancient environmental DNA

Group members
Current |
|||
|---|---|---|---|
| Name | Title | Co-supervised | |
| Christopher Bengt | PhD student, British Geological Survey | with Andrew Smith, British Geological Survey and Peter Wynn, Lancaster University | |
| Sara Behnamian | Postdoc | ||
| Dominik Noršić | PhD student | with Karina Krarup Sand | |
| Aikaterini Kafetzidou | Staff Scientist | ||
| Yike Huang | Postdoc | ||
| Alessandro Mereghetti | Postdoc | ||
| Swati Nielsen | PhD student | with Fernando Racimo | |
| Lasse Frøslev Hoppe | MSc student | ||
| Maja Seier | BSc student | ||
Alumni |
|||
| Milena Thiel | Msc student | With Nicole Posth | |
| Ivona Banicek | Visiting PhD student, University of Zagreb | ||
| Oluwatoosin Agbaje | Postdoc | with Karina K. Sand | |
| Pavel Ankon | Visiting PhD student, University of Zagreb |

